On January 15, 2025, a research team led by Baobin Li from the Institute for Translational Brain Research, Fudan University, in collaboration with Qingfeng Chen's team from the Center for Life Sciences at Yunnan University and Jean-luc Battini's team from the University of Montpellier in France, published a research paper titled "Structural basis of phosphate export by human XPR1" in Nature Communications. This paper reports the high-resolution cryo-EM structures of the human phosphate (Pi) export protein XPR1 in two distinct conformations. By combining in vitro transport assays, computational biology, and other methods, the study reveals the structural basis for XPR1-mediated Pi export to the extracellular space. This work has laid the groundwork for future in-depth mechanistic investigations and the development of therapeutics targeting this protein.
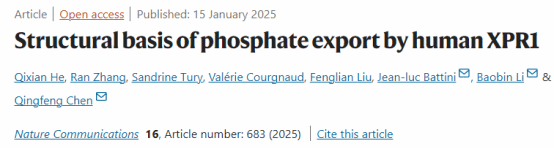
Phosphorus is essential for all living organisms and is crucial for maintaining life activities. At the cellular level, the uptake and export of inorganic phosphate (Pi) are mediated by specific transporters, which work together to maintain intracellular Pi homeostasis. Imbalances in intracellular Pi concentration can lead to a range of human diseases, such as rickets, cardiovascular diseases, and osteomalacia. In vertebrates, members of protein families like SLC20 and SLC34 are responsible for importing Pi into cells. In contrast, proteins involved in cellular Pi export are less well understood; XPR1 and its homologs are currently the only proteins reported to participate in Pi export. Dysfunction of XPR1 is associated with diseases such as primary familial brain calcification (PFBC) and thrombosis, and it can also serve as a diagnostic biomarker for tongue squamous cell carcinoma.
XPR1 consists of a transmembrane domain with multiple helical segments and an intracellular SPX domain located in the cytoplasm. Studies indicate that the SPX domain can sense intracellular inositol pyrophosphate (PP-InsP) levels, thereby regulating cellular Pi uptake, storage, and utilization. Apart from the crystal structure of the isolated SPX domain, structural information about XPR1 itself has remained scarce. Furthermore, whether XPR1 truly functions as a Pi export protein and the mechanism by which it exports Pi have remained open questions.
This study resolved the cryo-electron microscopy (cryo-EM) structures of the human XPR1 wild-type (XPR1WT) and a mutant defective in PP-InsP binding (XPR13mut), revealing the overall architecture of its homodimeric assembly. These structures feature a high-resolution transmembrane domain (TMD), while the SPX domain is largely invisible due to its inherent flexibility. Within the TMD, each monomer consists of 10 transmembrane helices (TMs). TM1, TM3, and TM4 form the scaffold domain, whereas TM2 and TM5-TM10 constitute the core domain. This core domain exhibits distant homology to ion-transporting rhodopsins.
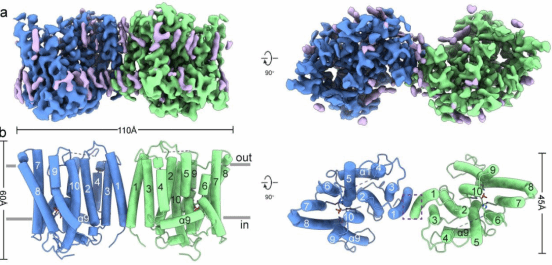
Within the core domain, the transmembrane helices (TMs) form a barrel-like architecture, creating a continuous ion translocation pathway. At the narrowest point of this pathway, located near the cytoplasmic side at its center, a phosphate (Pi) binding site is formed. This site effectively divides the pathway into an intracellular vestibule (IV) and an extracellular vestibule (EV). Studies indicate that impaired PP-InsP synthesis correlates with reduced Pi export levels in animal cells, suggesting that XPR1 activity may be activated by PP-InsP binding. Consistent with this hypothesis, structural analysis reveals that the extracellular vestibule (EV) in the PP-InsP binding site mutant XPR13mut is in a closed state, whereas the EV in the wild-type XPR1WT is in an open state. This conformational change is primarily due to structural rearrangements in TM9 and the side chain of residue W573. Collectively, these structural findings suggest that XPR1 functions via a mechanism more akin to an ion channel.
Qixian He (Master's candidate, School of Life Sciences, Yunnan University), Ran Zhang (Associate Research Fellow, Institute for Translational Brain Research, Fudan University), and Sandrine Tury (Institute of Infectious Diseases in Montpellier) are the co-first authors of the paper. Professor Qingfeng Chen (Yunnan University), Researcher Baobin Li (Institute for Translational Brain Research, Fudan University), and Professor Jean-luc Battini (University of Montpellier) are the corresponding authors.This research was supported by grants from the National Science and Technology Innovation 2030 Major Project of China, the National Natural Science Foundation of China (NSFC), the Fondation pour la Recherche Médicale (FRM), and other funding agencies.
Full article link: https://www.nature.com/articles/s41467-025-55995-8